Research
Nonhost Resistance Mechanism for Guaranteeing Plant Survival
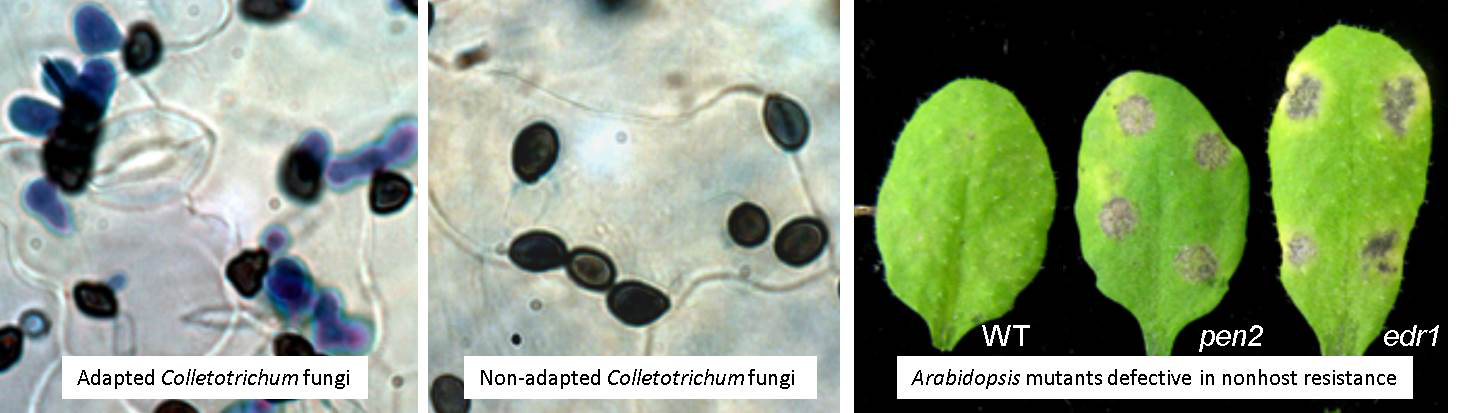
Phytopathogenic fungi belonging to the genus Colletotrichum, which cause anthracnose disease, generally infect a narrow range of plants, thus exhibiting stringent host specificity. For example, Colletotrichum orbiculare causes severe diseases in cucurbitaceae plants, but never infects the brassicaceae plant Arabidopsis thaliana. At this time, A. thaliana exhibits durable resistance, called “nonhost resistance,” to C. orbiculare. The molecular mechanism underlying nonhost resistance remains poorly understood; thus, our challenge is to unravel this mechanism using multifaceted analyses, including molecular genetic approaches. To date, we have identified regulatory factors and antifungal pathways that are critical for nonhost resistance and have found that a form of this defense system is multilayered, which confers robustness to nonhost resistance.
Counterstrategies of Plant Pathogens to Plant Resistance
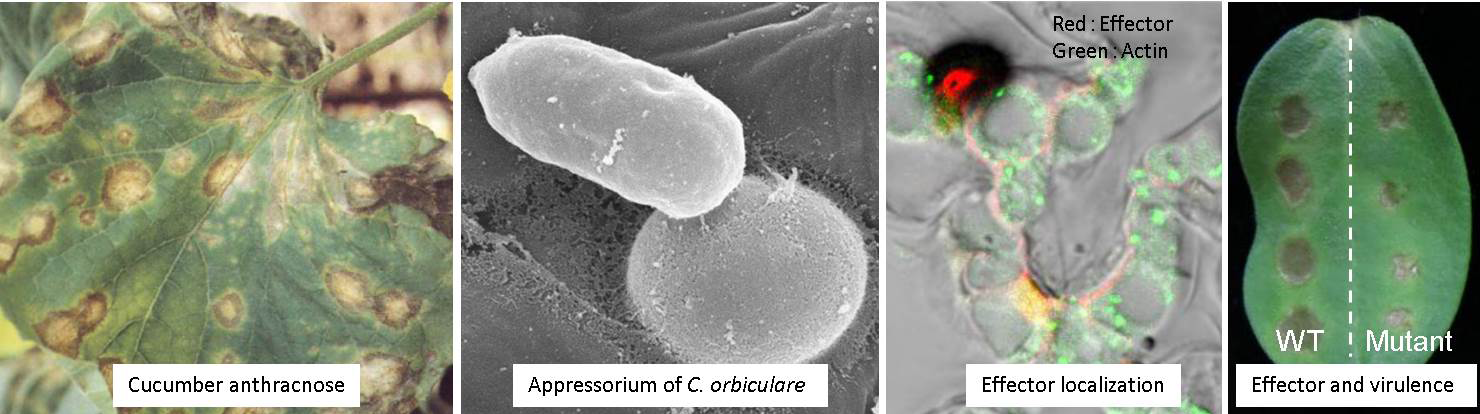
The robustness of nonhost resistance raises an intrinsic question: how do adapted pathogenic microorganisms evade or suppress nonhost resistance? For example, C. orbiculare must suppress the “nonhost resistance” of cucurbitaceae plants, and it is plausible that the pathogen deploys secreted molecules called effectors for the suppression of plant defenses. We are intensively studying Colletorichum effectors via cell biological analyses, as well as genome and transcriptome analyses. We aim to unravel the molecular background of the establishment of host specificity of plant pathogenic fungi via the study of effectors. In parallel, we are studying the interactions between plants and pathogenic microorganisms, such as fungi and bacteria, in the context of battles for nutrient acquisition.
Deciphering molecular interactions between plants and pathogens in diverse environments
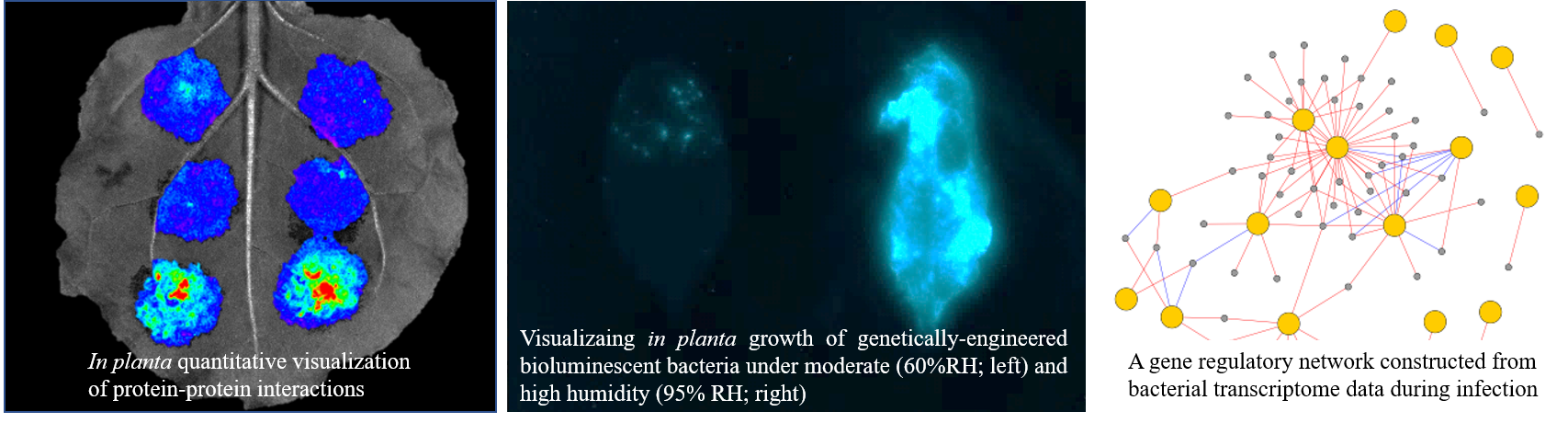
In nature, plants and microbial pathogens continuously fight against each other under ever-changing environments. The outcome of plant-pathogen interactions (e.g. disease development) is known to be highly influenced by environmental factors such as temperature and humidity. Yet, the mechanism behind this biological phenomenon is poorly understood. It is absolutely necessary to elucidate this mechanism toward controlling plant diseases under global climate change. We combine cutting-edge techniques in plant molecular genetics and molecular microbiology with bioinformatics such as functional genomics and network analysis, in order to decipher how high temperature and high humidity influence the plant and the bacterial pathogen during an active interaction.
Plant Resistance Mechanism to Plant RNA Viruses
Bromoviruses, which are a group of plant RNA viruses, show distinct host specificity. For example, the Brome mosaic virus (BMV) infects barley, but not A. thaliana, because of the latter’s nonhost resistance against BMV. In plants, the nonhost resistance against viruses remains poorly understood. We have analyzed the involvement of RNA silencing and unknown defense mechanisms in such nonhost resistance against viruses. We have also been attempting to isolate novel resistance (R) genes that are effective against bromovirus infection and to identify and analyze the function of bromoviral suppressors of RNA silencing.

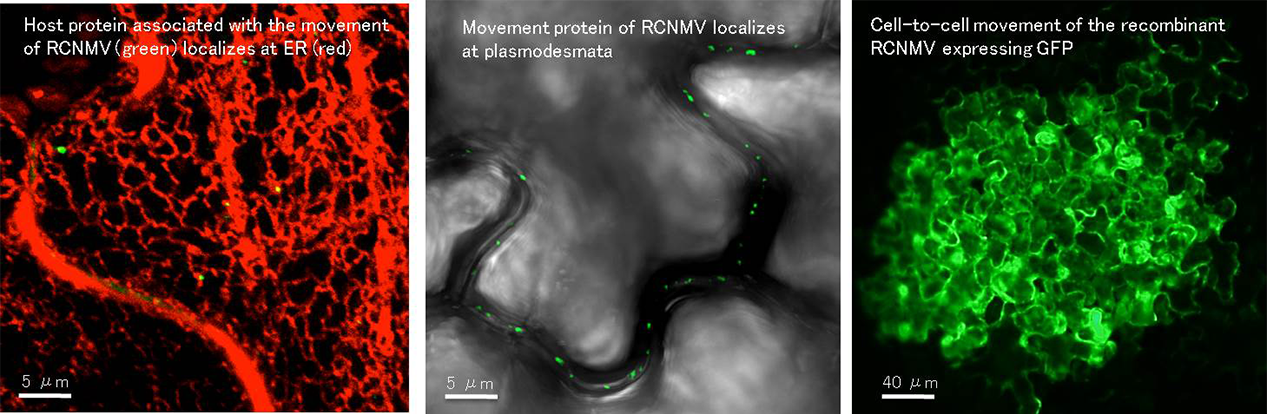
Cell-to-Cell Movement Mechanism of Plant RNA Viruses
Plant viruses encode a movement protein (MP) that facilitates viral cell-to-cell and long-distance movement through vasculature, which establishes the systemic infection of viruses. MP transports viral genomes to the plasmodesmata (PD), which are cytoplasmic conduits that connect plant cells, and opens the PD gates to convey viral genomes to neighboring cells. We are intensively studying the movement mechanism of Dianthoviruses, such as the Red clover necrotic mosaic virus and Carnation ringspot virus, which are positive-strand RNA viruses that infect legumes. We aim to unravel the molecular mechanisms of virus movement using structural and functional analyses of MPs and the identification and functional analyses of the host factor proteins that are involved in the viral movement processes.